Key points
- DISGENET has genetic support for 90% of FDA drug approvals in 2021
- The information provided by DISGENET surpasses publicly available resources, which can only support 66% of drug approvals
- DISGENET also offers information on disease biomarkers and different metrics to prioritize relevant targets for a wide array of indications
Human genetics and FDA drug approvals in 2021
There is compelling evidence of the value of incorporating genetic information in the drug development process. A drug target supported by genetic evidence has a 2-fold higher probability of successful clinical development compared to those targets with no genetic support [1]. A systematic analysis of FDA drug approvals showed that drugs with human genetics support were 2-5 fold more likely to lead to an improved therapy [2].
What is the genetic support for the 50 drugs that were approved by the FDA the last year?
What is the genetic support for the 50 drugs that were approved by the FDA the last year [3]? To answer this question requires accessing all current knowledge on the association of the drug target to the disease and data analytics capabilities. Knowledge platforms such as Open Targets and DISGENET facilitate accessing a vast array of evidence supporting the validity of targets for different therapeutic indications. A recent publication based on Open Targets estimates that for two-thirds of last year’s drug approvals (33 out of 50) the targets of the drugs, or a protein in the vicinity of the human interactome, have been associated with the indication, or a similar phenotype [4].
But, is it possible that this number is higher? Let’s take a closer look at the details and use DISGENET to assess whether the targets of the drugs have been previously reported as involved in the disease mechanism.
But before we start, is it fair to include in the analysis four drugs that have been developed for other organisms? This is the case for the combination of Cabotegravir and rilpivirine (Cabenuva Kit), two HIV-1 antiretroviral drugs, or Maribavir, an antiviral against human cytomegalovirus, Ibrexafungerp, an antifungal agent and Fexinidazole, an antiprotozoal used for African trypanosomiasis.
Three new drugs do not target a gene or protein. Melphalan flufenamide, indicated for Multiple myeloma, exerts its antitumor activity through DNA crosslinking [5]. This drug, approved in February 2021 was withdrawn later in the year [5]. Fosdenopterin, indicated for molybdenum cofactor deficiency (MoCD) Type A, is an exogenous form of cPMP, replacing endogenous production and allowing for the synthesis of molybdenum cofactor to proceed [6]. Another approval with a non-human target is the Asparaginase Erwinia chrysanthemi, an enzyme used to treat patients with acute lymphoblastic leukaemia and lymphoblastic lymphoma [7]. Asparagine is essential for the survival of leukemic cells, however, they lack the asparagine synthetase enzyme and depend on an exogenous source of asparagine. Asparaginase Erwinia chrysanthemi depletes the source of asparagine for leukemic cells [8]. Thus, in FDA approvals for 2021 only 43 drugs have at least one human protein target (Table 1).
Show Table 1: Drugs approved in 2021 with human targets.
The 50 approved drugs in 2021 treat 48 indications or diseases. Note that Nextstellis, a combination of Drospirenone, a spironolactone analogue, and estetrol, a synthetic analogue of a native estrogen, have been approved to prevent pregnancy, is not actually a disorder. Additionally, two imaging agents are listed in last year’s approvals: Pafolacianine for the detection of ovarian cancer and Piflufolastat F-18 for prostate cancer, respectively, by the use of molecular biomarkers.
We converted 39 indications to 44 Unified Medical Language System (UMLS) Concept Unique Identifiers (CUIs), the disease standard used in DISGENET using our mapping service. The publication that performed the original analysis provided the mappings to the Experimental Factor Ontology [9]. Using UMLS CUIs allowed us to map in a more exact fashion the indications for the drugs, due to the richer and more granular descriptions of phenotypes provided by the UMLS. For example, Severe hypoglycaemia can be mapped to the concept “Severe hypoglycaemia” (UMLS C4728082), while using the EFO a related concept such as “Diabetes mellitus” (EFO EFO 0001360) has to be chosen. Another example is Atopic dermatitis, which can be mapped to “Dermatitis, Atopic” (UMLS C0011615), instead of “atopic eczema” (EFO EFO_0000274). See table 2 for more details on the terms with improved mappings thanks to the use of the UMLS.
Show Table 2: The UMLS provides more accurate disease mappings than the EFO.
Sixty-two percent (31 out of 43) of the drugs have at least one target that is associated with the indication when considering the exact matching of the indication (Figure 1). Interestingly, DISGENET not only provides a comprehensive list of genes associated with the indications but offers metrics such as the DISGENET score to prioritize those genes. Some drugs specifically target the gene causing the disease, this is the case of APP for Alzheimer’s Disease, DMD for Duchenne Muscular Dystrophy, and GAA for Pompe disease. These genes are among the highest-scoring genes for their respective indications.
Other high-scoring genes are drivers of malignancies, such as KRAS, EGFR and MET, that are targets of 3 drugs to treat NSCLC. Other interesting cases are Pafolacianine, an imaging agent that targets the folate receptor alpha (FRα), which is often overexpressed in various cancers including ovarian cancer [10] and Piflufolastat F-18, which binds to the prostate-specific membrane antigen (PSMA), coded by FOLH1, which is higher in prostate cancer tissue and allows for the visualization of cancerous prostate tissue [11]. The relationship between FOLH1 and Prostatic Neoplasms has a score of 0.96 and over 600 publications support it. FOLR1 is associated with Malignant neoplasm of ovary with a score of 0.4 and 31 published studies.
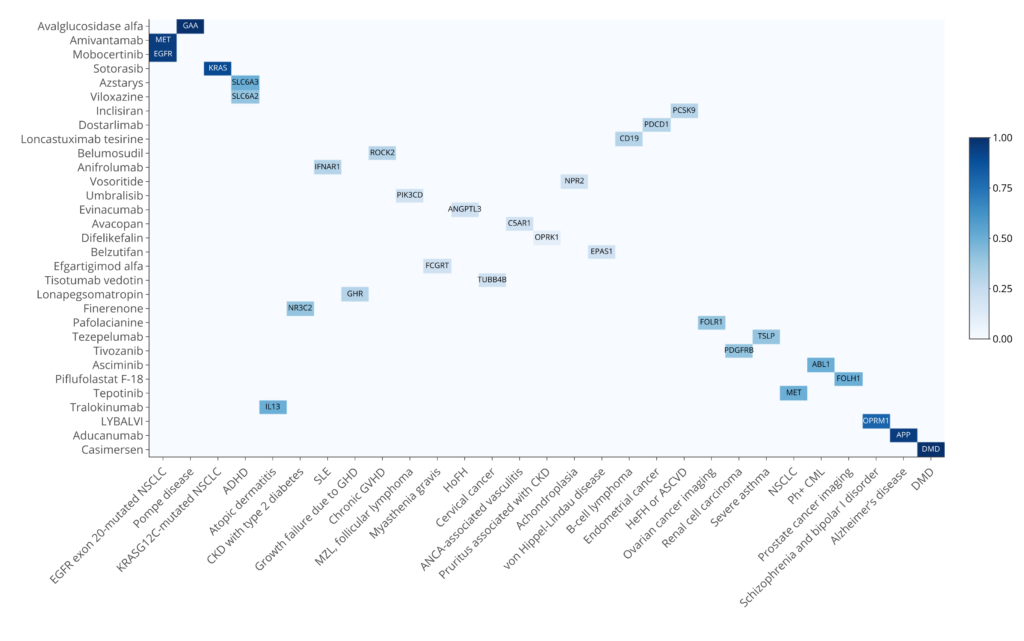
Figure 1: Drug-indications associations with genetic support in DISGENET. Exact mappings for the indications are considered. Note that for some drug-indication associations, there is more than one target with genetic support for the association with the indication. For these cases, we show the gene with the highest DISGENET score.
By expanding the disease mappings to similar diseases (Table 3), we find supporting evidence for 8 additional drugs. Therefore, 39 out of 43 drugs (90%) have evidence in DISGENET supporting a direct relationship between the target of the drug and the indication.
Table 3: Drugs that have genetic evidence for a similar phenotype of the indication
What are the four drugs with no genetic support? (Table 4). One of them is Infigratinib, prescribed for FGFR2-mutated bile duct cancer. There are around 1,000 associations in DISGENET relating FGF receptors 1-4 to multiple types of neoplasms, but not specifically to FGFR2-mutated bile duct cancer. Polycythaemia vera is associated with IFNA2, IFNA1, which are the ligands of the targets of Besremi, the drug approved for the disease. While Paroxysmal nocturnal hemoglobinuria (PNH) has been associated with C5 and other members of the complement cascade (information available in DISGENET), another member of the complement cascade has been selected as a target for Empaveli. This illustrates how in some cases due to different factors inherent in the drug discovery and design processes, such as the druggability of potential targets and other pharmaceutical considerations, the final target can be a protein different from the one initially characterized as involved in the disease mechanism, but otherwise belonging to the disease pathway.
Table 4: Drugs with no genetic support for their targets
For more information, a detailed analysis is available at:
disgenetplus_v20_FDA_drug_approvalsDownload
DISGENET: 90% Genetic Coverage for 2021 FDA Drug Approvals
In summary, this analysis shows that DISGENET has genetic support for 90% of the drugs approved during 2021 by the FDA. This contrasts with previous reports using publicly available tools, which achieved 66% of genetic support for drug approvals [4]. Thus, DISGENET is a key resource for drug R&D to provide actionable information on potential targets for a wide range of indications.
In addition, it illustrates the potential of DISGENET as a resource for information on disease biomarkers, as it also contains the evidence that relates the association of the biomarkers with their indications for the two imaging agents approved by the FDA.
References
[1] Cook D, Brown D, Alexander R, March R, Morgan P, Satterthwaite G, et al. Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework. Nat Rev Drug Discov 2014 136 2014;13:419–31. https://doi.org/10.1038/nrd4309.
[2] Nelson MR, Johnson T, Warren L, Hughes AR, Chissoe SL, Xu CF, et al. The genetics of drug efficacy: opportunities and challenges. Nat Rev Genet 2016 174 2016;17:197–206. https://doi.org/10.1038/nrg.2016.12.
[3] Mullard A. 2021 FDA approvals. Nat Rev Drug Discov 2022;21:83–8. https://doi.org/10.1038/D41573-022-00001-9.
[4] Ochoa D, Karim M, Ghoussaini Maya, Hulcoop DG, McDonagh EM, Dunham I. Human genetics evidence supports two-thirds of the 2021 FDA-approved drugs. Nat Rev Drug Discov 2022. https://doi.org/10.1038/D41573-022-00120-3.
[5] Olivier T, Prasad V. The approval and withdrawal of melphalan flufenamide (melflufen): Implications for the state of the FDA. Transl Oncol 2022;18:101374. https://doi.org/10.1016/J.TRANON.2022.101374.
[6] Farrell S, Karp J, Hager R, Wang Y, Adeniyi O, Wang J, et al. Regulatory news: Nulibry (fosdenopterin) approved to reduce the risk of mortality in patients with molybdenum cofactor deficiency type A: FDA approval summary. J Inherit Metab Dis 2021;44:1085–7. https://doi.org/10.1002/JIMD.12421.
[7] Maese L, Rau RE. Current Use of Asparaginase in Acute Lymphoblastic Leukemia/Lymphoblastic Lymphoma. Front Pediatr 2022;10:1030. https://doi.org/10.3389/FPED.2022.902117/BIBTEX.
[8] Fda. ERWINAZE (asparaginase Erwinia chrysanthemi). n.d.
[9] Malone J, Holloway E, Adamusiak T, Kapushesky M, Zheng J, Kolesnikov N, et al. Modeling sample variables with an Experimental Factor Ontology. Bioinformatics 2010;26:1112–8. https://doi.org/10.1093/BIOINFORMATICS/BTQ099.
[10] FDA approves pafolacianine for identifying malignant ovarian cancer lesions | FDA n.d. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-pafolacianine-identifying-malignant-ovarian-cancer-lesions (accessed October 4, 2022).
[11] Keam SJ. Piflufolastat F 18: Diagnostic First Approval. Mol Diagnosis Ther 2021 255 2021;25:647–56. https://doi.org/10.1007/S40291-021-00548-0.
