In recent years, there has been an increasing appreciation of the need to consider variability in human populations in evaluating health risks for new and existing chemicals. Recent policies such as The Frank R. Lautenberg Chemical Safety for the twenty-first Century Act (2016) require the US Environmental Protection Agency to evaluate new and existing toxic chemicals with explicit consideration of susceptible populations of all types (life stage, exposure, genetic, etc.). One way to achieve this goal is to identify individuals who are more susceptible to suffering from adverse health effects upon exposure to a chemical because of their genetic background.
Research and developments in the area of genomics have resulted in increased availability of large-scale data sets on human genomic variability across different populations [1,2] including the assessment of the effect of genomic variability on disease risk [3–5]. There is an untapped opportunity to exploit this wealth of data to incorporate information on human variability at the genomic level to inform chemical risk assessment and improve the estimation of risk for specific human populations. For instance, there are 200 studies on environmental exposures registered in the GWAS Catalog (accessed on January 16th, 2023), which could be leveraged to support the development of methods to anticipate the toxicity of chemicals in humans.
The use of genetic information to support safety assessment in the development of therapeutic drugs is more advanced. Recently, Carss and colleagues reviewed the different data resources and available methods to leverage human genetic data to support the safety assessment of drugs [8]. Information on human genetics is frequently used in drug discovery pipelines to support the identification of potential drug targets based on their relevance to disease in humans. There is an increasing interest in using genetic data to identify potential safety liabilities of modulating a given target. It has been proposed that human genetic variants can be used as a model to anticipate the effect of lifelong modulation of therapeutic targets and identify the potential risk for on-target adverse events. Off-target toxicities could also be predicted based on current knowledge of disease-associated genes and genomic variants. Genetic-based approaches for safety evaluation of novel therapeutics at pre-clinical stages are particularly appealing for those compounds that lack pharmacologically relevant animal models and therefore can contribute to the intrinsic safety profile of a drug target. Moreover, genetic-based approaches can contribute to reducing the number of animals used in research.
One of the genomics resources proposed by Carss and colleagues to support drug safety assessment is DisGeNET. Its successor, DISGENET, contains a comprehensive and up-to-date catalogue of genes and genomic variants associated with human diseases and their phenotypic manifestations. As such, DISGENET is a key resource to support the identification of safety liabilities that might result from modulating targets by drugs from a wide spectrum of modalities. Moreover, genetic-informed approaches that are more and more used in the pharmaceutical industry could also be applied to the evaluation of the safety liabilities of chemicals in humans.
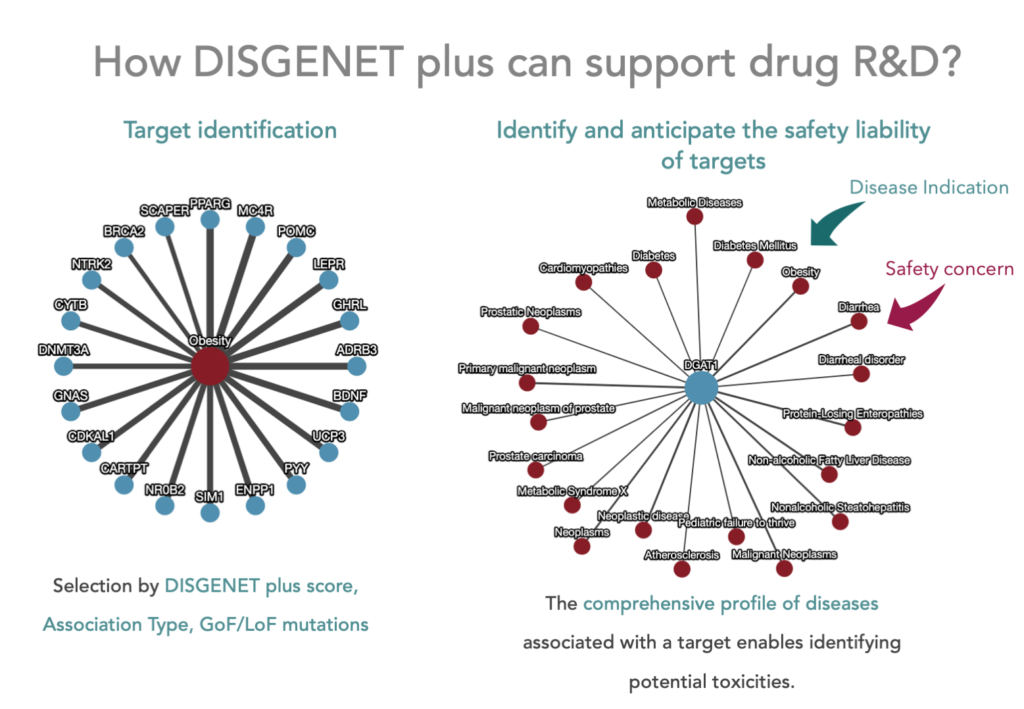
In this regard, there is an increasing interest in using the Adverse Outcome Pathway (AOP) framework to organise, identify, and characterise human genetic susceptibility in human chemical risk assessment [6]. An Adverse Outcome Pathway is a structured representation of biological events leading to adverse effects on health and is widely used for chemical risk assessment. The AOP links in a linear way existing knowledge along one or more series of causally connected key events (KE) between two points — a molecular initiating event (MIE) and an adverse outcome (AO) that occur at a level of biological organization relevant to risk assessment. The linkage between the events is described by key event relationships (KER) that describe the causal relationships between the key events. A recent report analysed publicly available human genetic datasets and assessed how they could be leveraged to gain a mechanistic understanding of molecular events and therefore characterise human susceptibility to an AO [6]. A computational approach to prioritize AOPs with potential for human genetic variability, based on mapping MIE, KE and AO to human genes and characterising their genomic variability was proposed. This AOP-anchored genetic susceptibility approach integrates information on disease-associated genes and genomic variants, population-based allele frequency information for the genomic variants, as well as data on the functional impact of the genomic variants derived from high-throughput screening and/or in silico approaches. The authors suggest a general approach based on the integration of two data sources: mechanistic data implicating chemical-gene targets and genomics data characterising genes implicated in pathways relevant to chemical toxicity [7]. Information on the association of genes and proteins to diseases and adverse phenotypes from DISGENET could be incorporated into AOPs to support the use of human genomic variability in chemical risk assessment. In the context of the RISKHUNT3R project, an European effort to develop a new modular framework for animal-free next-generation chemical risk assessment (NGRA), we are developing new approaches for incorporating human genetics in NGRA.
If you want to learn more about how DISGENET can support risk assessment of chemicals or target de-risking, contact us at info@disgenet.com.
References
[1] Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–9. https://doi.org/10.1038/s41586-018-0579-z.
[2] Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–43. https://doi.org/10.1038/s41586-020-2308-7.
[3] Lappalainen T, MacArthur DG. From variant to function in human disease genetics. Science (80-) 2021;373:1464–8. https://doi.org/10.1126/science.abi8207.
[4] Giacomini KM, Yee SW, Mushiroda T, Weinshilboum RM, Ratain MJ, Kubo M. Genome-wide association studies of drug response and toxicity: an opportunity for genome medicine. Nat Rev Drug Discov 2017;16:70–70. https://doi.org/10.1038/nrd.2016.234.
[5] Uffelmann E, Huang QQ, Munung NS, de Vries J, Okada Y, Martin AR, et al. Genome-wide association studies. Nat Rev Methods Prim 2021 11 2021;1:1–21. https://doi.org/10.1038/s43586-021-00056-9.
[6] Mortensen HM, Chamberlin J, Joubert B, Angrish M, Sipes N, Lee JS, et al. Leveraging human genetic and adverse outcome pathway (AOP) data to inform susceptibility in human health risk assessment. Mamm Genome 2018:1–15. https://doi.org/10.1007/s00335-018-9738-7.
[7] Mortensen HM, Euling SY. Integrating mechanistic and polymorphism data to characterize human genetic susceptibility for environmental chemical risk assessment in the 21st century. Toxicol Appl Pharmacol 2013;271:395–404. https://doi.org/10.1016/j.taap.2011.01.015.
[8] Carss KJ, Deaton AM, Del Rio-Espinola A, Diogo D, Fielden M, Kulkarni DA, et al. Using human genetics to improve safety assessment of therapeutics. Nat Rev Drug Discov 2022. https://doi.org/10.1038/s41573-022-00561-w.