Key points
- Flupirtine, a non-opioid analgesic introduced in 1984, took a tumultuous journey due to concerns over liver toxicity, despite initial approval for pain management.
- The DISGENET data analysis platform proves crucial in unveiling the complex connections between drug-associated genes and liver conditions, potentially predicting adverse reactions.
- Genes influenced by flupirtine exhibit substantial links to liver diseases, with insights provided by DISGENET.
- By harnessing DISGENET, it might have been possible to anticipate toxicity concerns in early drug development and implement vigilant monitoring during clinical trials and post-approval phases, avoiding unforeseen medical consequences.
- Looking beyond flupirtine, the story highlights the essential role of data analysis platforms like DISGENET in anticipating adverse drug reactions and guiding safer drug development.
Flupirtine, an aminopyridine introduced in 1984 as a non-opioid analgesic for acute and chronic pain, embarked on a tumultuous journey within the medical landscape. Initially approved for broad pain management, its trajectory shifted in 2013 due to concerns over liver toxicity. The European Medicines Agency (EMA) limited its use to acute pain for a maximum of two weeks and exclusively for individuals unsuited to other painkillers. However, these precautions yielded only short-lived benefits. By March 2018, the EMA had recommended the withdrawal of marketing authorizations, indicative of persisting instances of severe liver injuries, including liver failure, despite imposed restrictions.
Could the emergence of liver toxicity concerns from flupirtine treatment have been anticipated through the DISGENET platform?
The drug’s primary mechanism of action involves indirect NDMA receptor antagonism by activating potassium channels. It represents the inaugural member of the ‘selective neuronal potassium channel openers’ pharmacological class. Specifically, it operates by selectively opening potassium channels, including those encoded by the KCNQ2 and KCNJ3 genes. However, as with many drugs, flupirtine’s scope of action extends to other proteins as well. By querying public resources like PubChem, DrugBank, Chembl, and PharmGKB for information on flupirtine-related targets and genes, we compiled a comprehensive list of 21 genes associated with flupirtine.
Analyzing the disease profiles of these flupirtine-associated genes might uncover indicators of liver toxicity. In this pursuit, DISGENET emerges as a valuable tool, streamlining access to pertinent information. Using DISGENET is equivalent to reading over 30M scientific publications and extracting the information relevant to diseases and phenotypes associated with the drug targets in a single click.
All the genes associated with flupirtine exhibit links to disease phenotypes within DISGENET, contributing to over 3,800 gene-disease associations.
Significantly, DISGENET findings corroborate substantial evidence linking flupirtine-affected genes to liver diseases and related traits. Most of the genes directly influenced by flupirtine (17 from 21 genes) correlate with liver conditions. Notably exempt from this association are genes encoding potassium channels such as KCNJ3, KCNJ5, KCNJ6, and KCNQ2. In contrast, genes like ADRA2A, BCL2, and VDR are implicated in liver-related conditions. The central figure here is the vitamin D receptor (VDR), which plays a pivotal role in processing the biological effects of vitamin D—a vital fat-soluble nutrient governing bone health, immune function, and mineral regulation. With a presence in various tissues, including the liver, the VDR has been extensively studied in relation to liver-related disorders like NAFLD, NASH, viral hepatitis, liver fibrosis, and liver cancer, among others. Figure 1 displays the gene-disease associations of the top-scoring associations of flupirtine genes and liver conditions. Table 1 provides detailed information on the liver conditions associated with flupirtine genes, along with the DISGENET score and the first and last year evidence of the disease-association have been published.
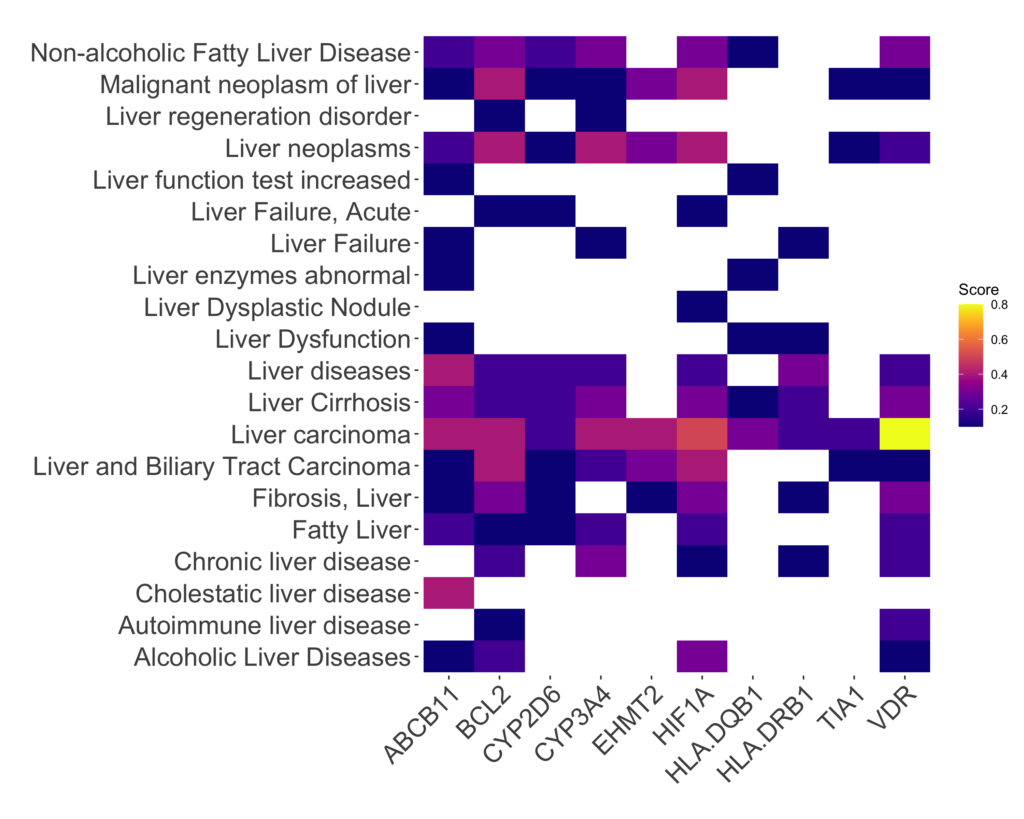
Noteworthy is the fact that some of these connections were established prior to the EMA’s 2013 restrictions (see Table 1) . By harnessing DISGENET, it might have been possible to pre-emptively shed light on potential adverse reactions to the drug. Incorporating this information could have facilitated early identification of safety signals during pre-clinical phases, or vigilant monitoring of liver events during clinical trials and post-approval phases via pharmacovigilance channels. This underscores the pivotal role of comprehensive data analysis platforms like DISGENET in shaping drug development and regulatory decisions, potentially averting unforeseen medical consequences.
Show Table 1: Liver Diseases associated with Flupirtine genes
| Gene | Disease | DISGENET Score | First year | Last year |
|---|---|---|---|---|
| ABCB11 | Cholestasis, Progressive Familial Intrahepatic, 2 | 0.9 | 1993 | 2022 |
| ABCB11 | Intrahepatic Cholestasis | 0.8 | 2002 | 2021 |
| ABCB11 | Progressive intrahepatic cholestasis (disorder) | 0.8 | 1998 | 2022 |
| ABCB11 | Cholestasis, benign recurrent intrahepatic 2 | 0.7 | 1993 | 2021 |
| ABCB11 | Cholestasis, progressive familial intrahepatic 3 | 0.6 | 2006 | 2010 |
But we can still go one step beyond: Do specific individuals possess a heightened susceptibility to flupirtine-induced liver toxicity? Could this susceptibility stem, at least in part, from genomic variability? Can DISGENET provide such insights? If so, pharmacogenomic guidelines for the drug could be devised, allowing for more personalized treatment approaches. This way, non-susceptible patients could still benefit from flupirtine treatment. This is especially appealing in light of ongoing studies testing the effect of flupirtine for alleviating pain in different conditions and procedures, or evaluating its therapeutic efficacy for the management of Creutzfeldt-Jakob disease [1] or Multiple Sclerosis [2].
One prime example of a genomic variant linked to flupirtine toxicity is a variant within the HLA-DQB1 gene. A GWAs study revealed that the HLA-DQB1 *05:02 haplotype heightens the risk of drug-induced liver injury during flupirtine treatment [3].
However, numerous other variants within genes associated with flupirtine also intertwine with liver conditions. For instance, genomic variants in genes like VDR, BCL2, or ABCB11 contribute to this connection.
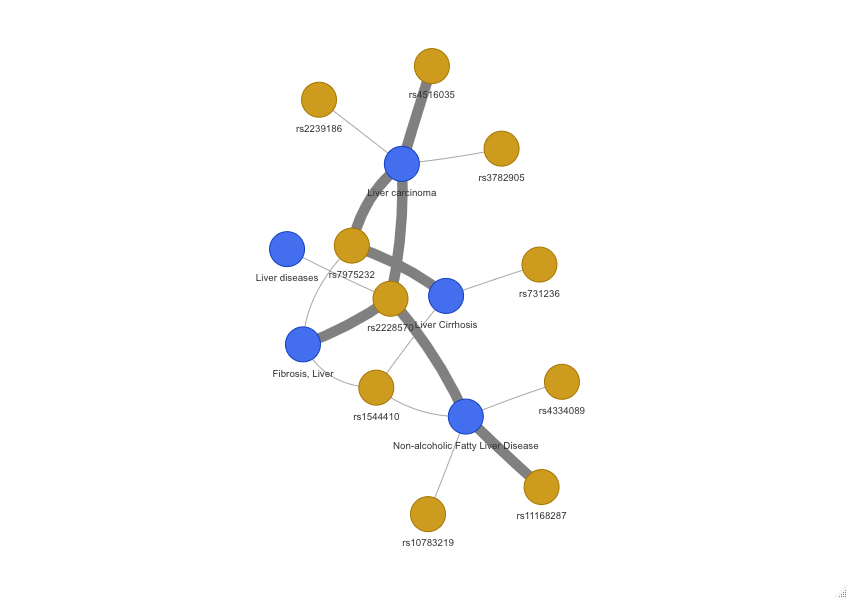
The VDR gene harbours dozen genomic variants tied to liver conditions. Figure 2 illustrates some of the associations among VDR SNPs and liver disorders. Take, for instance, the rs2228570 variant, which causes a loss of the start codon. This variant is prevalent among African/African American populations and South Asians (allele frequencies of 0.7868 and 0.7675, respectively), but less so among East Asians and Latinos (allele frequencies of 0.5423 and 0.5259, respectively). Another variant, rs1544410, exhibits marked differences in allele frequencies across populations. While it is observed at 0.4172 in Ashkenazi Jewish individuals, East Asians show notably lower frequency (0.04936). This variant is associated with liver fibrosis and NAFLD, potentially contributing to differential susceptibility to liver toxicity during flupirtine treatment.
Summary
Flupirtine, initially introduced as a non-opioid analgesic, encountered a turbulent journey due to liver toxicity concerns. Despite regulatory efforts, severe liver injuries persisted. DISGENET, a data analysis platform, offers insights into the complex relationship between flupirtine-associated genes and liver conditions. The central role of the vitamin D receptor in liver health is highlighted. By leveraging DISGENET, preemptive identification of adverse reactions becomes possible, shaping drug development and regulatory decisions. Furthermore, genomic variants, such as those within the VDR gene, might contribute to the intricate susceptibility to flupirtine-induced liver toxicity, underlining the potential for personalized treatment guidelines.
Conclusion
The story of flupirtine underscores the importance of data analysis platforms like DISGENET in anticipating adverse drug reactions. By dissecting genetic links between drugs and diseases, DISGENET can inform safer drug development. Flupirtine’s impact on liver health, and the genomic variants influencing susceptibility, exemplify the power of personalized medicine in minimizing unforeseen medical consequences.
If you want to learn more on how to leverage DISGENET for anticipating safety issues in drug development, contact us at info@disgenet.com
References
[1] Miranda, Luiz Henrique Lélis, et al. “Systematic review of pharmacological management in Creutzfeldt-Jakob disease: no options so far?.” Arquivos de Neuro-Psiquiatria 80 (2022): 837-844.
[2] Dörr, Jan, et al. “Disease Modification in Multiple Sclerosis by Flupirtine—Results of a Randomized Placebo Controlled Phase II Trial.” Frontiers in Neurology 9 (2018): 842.
[3] Nicoletti, Paola, et al. “HLA-DRB1* 16: 01-DQB1* 05: 02 is a novel genetic risk factor for flupirtine-induced liver injury.” Pharmacogenetics and genomics 26.5 (2016): 218-224.